“Genome editing” or “Genome engineering” gives the ability to introduce a variety of genetic alterations (deletion, insertion…) into mammalians cells. During the past decade, zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) were the tools of choice for genome editing technologies until the recent discovery of CRISPR/Cas9 technology that have revolutionised the field.
Successful CRISPR/Cas9 genome editing can be performed through diverse approaches (plasmids, mRNA, nuclease, viral delivery). Accordingly, efficient nucleic acid delivery (transfection or transduction) represents a critical step for genome editing experiments.
With more than 10 years of expertise in the development of transfection reagents, OZ Biosciences offers tailored transfection solutions for CRISPR/Cas9 technology.
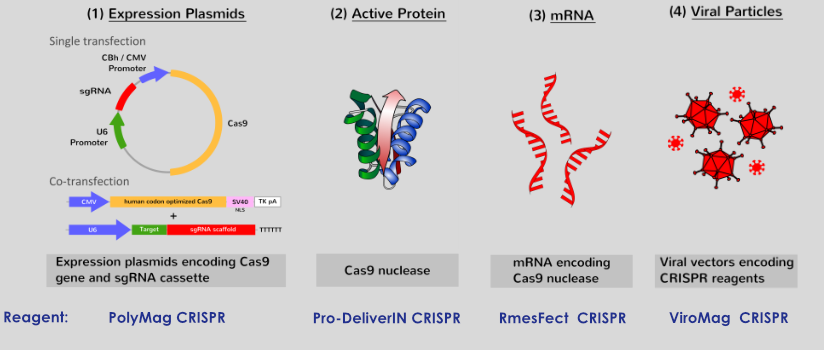
Types of Delivery System
To generate cellular models, Cas9 and the designed single guide RNA (sgRNA; a chimeric RNA containing all essential crRNA and tracRNA components) can be introduced into the target cells. The type II CRISPR/Cas system only needs a single Cas protein that can be expressed into target cells by: (1) plasmid transfection, (2) direct delivery of the active Cas9 endonuclease, (3) transfection of mRNA encoding for Cas9 or (4) by viral vectors transduction.
PolyMag CRISPR
PolyMag CRISPR is the only magnet assisted transfection reagent optimized to deliver high level of plasmid DNA and/or mRNA expressing Cas9 and guide RNA (gRNA). This reagent is based on our proprietary Magnetofection™ technology. The nanoparticles are composed of iron oxide magnetic core coated with cationic molecules. For your gene editing experiments, this reagent provides high transfection efficiency without toxicity.
Main features
- High-efficiency delivery of CRISPR/Cas9 system even on primary and hard-to-transfect cells.
- Ideal for co-transfection of pDNA/pDNA, pDNA/gRNA, pDNA/mRNA.
- Optimised for large plasmid DNA containing Cas9, and guide RNA.
- Low cell toxicity – enabling experiments with fewer cells.
Pro-DeliverIN CRISPR
Pro-DeliverINTM CRISPR is a transfection reagent optimized for recombinant Cas9 protein delivery or Cas9/gRNA RNP complexes. For your gene editing applications, this reagent provides high transfection efficiency with minimal toxicity. Why choose Cas9 protein instead of Cas9 DNA or mRNA? The Cas9 recombinant protein is delivered more rapidly than nucleic acids and is fully active once inside the cells without latency period (in contrast to transcription and translation machineries required for the nucleic acids).
Main features
- High-efficiency delivery of Cas9 protein or Cas9 gRNA complexes.
- Low cell toxicity – enabling experiments with fewer cells.
RmesFect CRISPR
RmesFect™ CRISPR transfection reagent based on the TEE-technology is specifically designed for mRNA/gRNA transfection with high efficiency and low toxicity. RmesFect™ is efficient in a large variety of cells.
Main features
- Highly efficient: achieve RNAs delivery in all cells.
- Ready-to-use: no need for additional buffer.
- Low nucleic acid amount: minimised toxicity.
- Protects RNAs against degradation.
- Compatible with any culture medium.
- Serum compatible: medium change not required.
ViroMag CRISPR
Need to enhance the transduction efficiency of your CRISPR/Cas9 recombinant virus? ViroMag CRISPR reagent is the only magnetic viral transduction enhancer for CRISPR/Cas viruses (adenovirus, lentivirus, retrovirus…) that infects cells for gene editing applications. This reagent demonstrates an exceptionally high efficiency to promote, control and assist viral transductions so that no molecular biology processes or biochemical modifications are required.
Main features
- Highly increased transduction efficiency of viral CRISPR/Cas9 system.
- Enables genome editing even in primary, hard-to-transduce and non-permissive cells.
- Accelerates the transduction process.
- Concentrate the viral dose onto the cells.
- Synchronize cells adsorption/infection without modifications of the viruses.
Contact us to know more about how these products could help you with your research or continue reading to know more about CRISPR/Cas9.
Genome Editing with CRISPR/Cas9
In 2013, four groups demonstrated that CRISPR/Cas9 associated with guide RNA can be used for gene editing. [1-4] Based on the type II CRISPR/Cas9 mechanism, researchers created a single guide RNA (sgRNA), which is able to bind to a specific dsDNA sequence. This resulted in double-strand breaks (DSB) at the target site with: (1) a 20-bp sequence matching the protospacer of the guide RNA and (2) a protospacer-adjacent motif (PAM) 3 bp downstream NGG sequence. CRISPR/Cas9-mediated genome editing thus depends on the generation of DSB and subsequent cellular DNA repair process, including non-homologous end-joining (NHEJ)-mediated error-prone DNA repair and homology-directed repair (HDR)-mediated error-free DNA repair. Insertions and deletion mutations at the target site generated by NHEJ and HDR allow disrupting or abolishing the function of a target gene. Moreover, modifications in this system can also be used to silence genes, insert new exogenous DNA or block RNA transcription.
How does CRISPR/Cas9 work?
CRISPR/Cas9 system originates from bacteria in which provides acquired immunity against invading foreign DNA via RNA-guided cleavage. [5] Bacteria collect “protospacers”, short segments of foreign DNA (e.g. from bacteriophages) and integrate them into their genome. Sequences from CRISPR genomic loci are then transcribed into short CRISPR RNA (crRNA) that anneal transactivating crRNA (tracrRNAs) to destroy any DNA sequence matching the protospacers. After transcription and processing, crRNA first complexes with Cas9 and tracrRNA and then bind its target sequence onto DNA. Both R-loop forms and DNA strands are cut. crRNA is used as a guide while Cas9 acts as an endonuclease to cleave the DNA (figure 1).
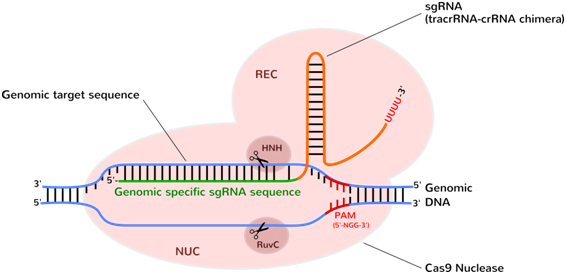
Figure 2: The CRISPR-Cas9 nuclease programmed with sgRNA. Upon binding the sgRNA guide (tracrRNA-crRNA) specifically targets a short DNA sequence-tag (PAM) and unzips DNA complementary to the sgRNA. sgRNA–target DNA heteroduplex, triggering R-loop formation results in a further structural rearrangement: Recognition (REC) and Nuclease lobes (NUC) undergo rotation to fully enclose the DNA target sequence. Two nuclease domains (RuvC, HNH) each nicking one DNA strand, generate a double-strand break. Structurally, REC domain interacts with the sgRNA, while NUC lobe drives interaction with the PAM and target DNA.
Various Cas9-based applications:
- Indel (insertion/deletion) mutations,
- Specific sequence insertion or replacement
- Large deletion or genomic rearrangement (inversion or translocation)
- Fusion to an activation domain:
- Gene Activation
- Other modifications (histone modification, DNA methylation, fluorescent protein)
- Imaging location of genomic locus.
CRISPR/Cas9 advantages over ZFNs and TALENs
CRISPR/Cas9 can easily be adapted to any genomic sequence by changing the 20-bp protospacer of the guide RNA; the Cas9 protein component remaining unchanged. This ease of use presents a main advantage over ZFNs and TALENs in generating genome-wide libraries or multiplexing guide RNA into the same cells.
- ZFNs and TALENs are built on protein-guided DNA cleavage that needs complex protein engineering.
- CRISPR/Cas9 only needs a short guide RNA for DNA targeting.
- CRISPR/Cas9 allows using several gRNA with different target sites: simultaneously genomic modifications at multiple independent sites. [1]
- Accelerates the generation of transgenic animals with multiple gene mutations. [6]
CRISPR/Cas9 system presents a versatile and reliable genome editing tool to facilitate a large variety of genome targeting applications. CRISPR/Cas9 components comprise an endonuclease and a sgRNA that can be delivered into cells under various forms (i.e. plasmid, mRNA, nuclease, virus).
References
- Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339 (6121):819-823.
- Mali P, et al. RNA-guided genome engineering via Cas9. Science. 2013;339 (6121):823-826.
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337 (6096):816-821.
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230-232.
- Wiedenheft B, et al. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331-338.
- Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153 (4):910-8.