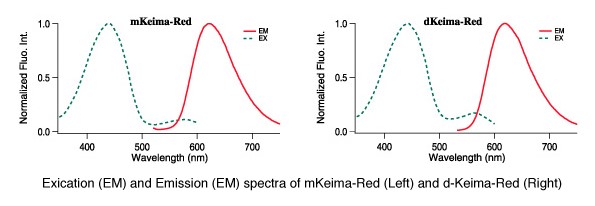
CoralHue® monomeric and dimeric Keima-Red are red fluorescent proteins with the largest commercially available Stokes shift (ex. 440 nm, em. 620 nm) making Keima-Red a superb reporter protein for multicolour fluorescence analysis. Because of this characteristic, they are excited by a very short wavelength but emit a long wavelength. Keima is named after a shogi (Japanese chess) piece Keima that can move in a hopping manner, similar to the knight in the game of chess.
The Keima-Red protein cloning plasmids allow for the insertion of cDNA sequences to create protein fusion products between the protein of interest and Keima. Keima-Red cloning plasmids create protein fusion products that are useful for tracking protein localisation within cells as well as monitoring gene expression. Keima-Red is also available as target-specific constructs which allow for Keima-Red protein fusion products to be directed to either the mitochondria or the plasma membrane.
Performance and Use
Labelling Organelles
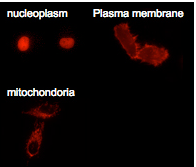
dKeima-Red and mKeima-Red are easily expressed in a wide range of organisms. dKeima-Red and mKeima-Red have been demonstrated in the nucleoplasm, plasma membrane, and mitochondria targeting signal models.
Multicolour Labelling of Mammalian Cells
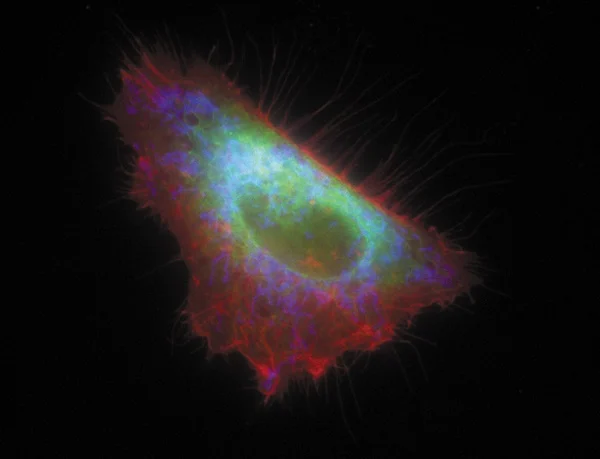
An image of the HeLa cell labelled with MBL fluorescent proteins targeting different organelles. mKO1 is localised on the plasma membrane (red), mAG1 in the endoplasmic reticulum (green) and mKeima-Red in the mitochondria (blue).
The image and information are provided by Dr. Miyawaki Atsushi, Laboratory for Cell Function and Dynamics, BSI, RIKEN.
Simultaneous four-color imaging of subcellular structures in a Vero cell using a single laser line (458 nm)
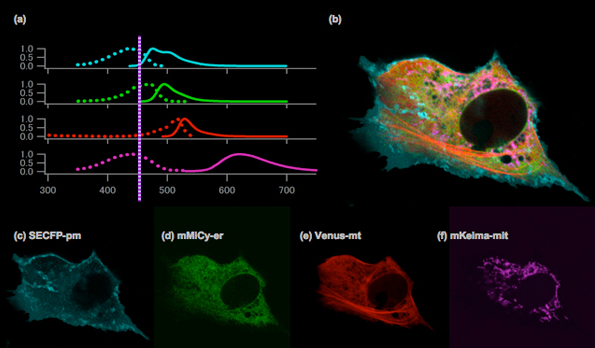
Normalised excitation and emission spectra of SECFP, mMiCy, Venus and mKeima (a). An image of the Vero cell with SECFP localised on the plasma membrane (cyan), mMiCy in the endoplasmic reticulum (green), YFP along the microtubules (red) and mKeima in the mitochondria (purple) (b). The image was created by merging the following images, which were obtained using spectral imaging: SECFP-pm (c), mMiCy-er (d), Venus-mt (e) and mKeima-mit (f).
Spectra imaging with a single laser line at 458 nm (Ar ion laser) was performed using the 32 channels of the LSM 510 META system (Carl Zeiss).
Images were kindly provided by Takako Kogure, Atsushi Miyawaki (Laboratory for Cell Function and Dynamics, BSI, RIKEN).
Mitophagy Detection
Keima-Red for mitophagy detection in living cells
- pH-dependent excitation profile
- Simple method for creating transfectant cells
- Use with any mammalian cells
- Useful for monitoring mitophagy
Mitophagy is the selective degradation of old or depolarised mitochondria by autophagy and contributes to maintaining a healthy population of mitochondria. Since damaged mitochondria lead to collapse cell homoeostasis, mitophagy is believed to be protective against diseases related to mitochondrial dysfunction such as in neurodegenerative disorders.
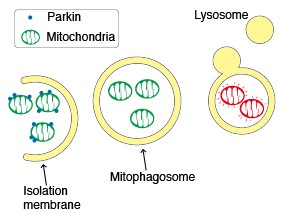
Parkin, an ubiquitin ligase known as the gene responsible for Parkinson’s disease, plays an important role in the autophagic elimination of mitophagy. When mitochondria are depolarised and dysfunctional, PTEN-induced putative kinase protein 1 (PINK1) accumulates on the outer membrane and recruits Parkin on the damaged mitochondria. The outer membrane on the mitochondria is then ubiquitinated through the ubiquitin ligase activity of Parkin. Finally, the poly-ubiquitinated mitochondria are selectively recognised and executed by the autophagic process.
Emission wavelength character of Keima-Red under various pH condition
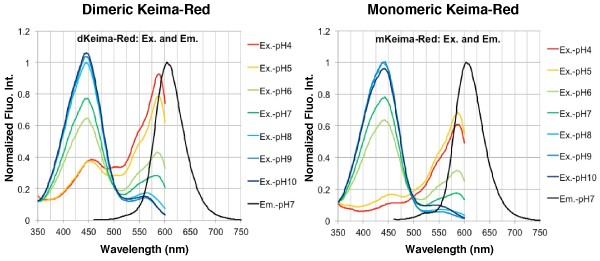
The fluorescent protein Keima has an excitation spectrum that changes according to pH. A short wavelength (440 nm) is predominant for excitation in a neutral environment, whereas a long wavelength (586 nm) is predominant in an acidic environment. The ratio of fluorescent intensity in each excitation condition is an indicator of mitophagy in living cells.
Mitochondrial targeting-mKeima-Red for mitophagy detection
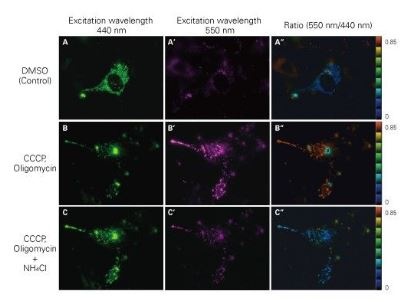
When MT-mKeima-Red, which is the Keima tagged with a mitochondrial-localised signal peptide sequence, and Parkin is expressed in target cells, mitophagy is detected and visualised through the difference in the fluorescent wavelengths observed before and after drug treatment. Mitophagy is induced by the administration of CCCP and oligomycin, drugs that affect mitochondrial membrane potential and is observed with 550nm/440nm ratio images (A”, B”, C”).
Through mitophagy induction, the mitochondria-localised MT-mKeima-Red is displayed in red in ratio images showing its localisation in an acidic environment (B-B”). The neutraliser NH4Cl is then administered which forces the entire cell into a neutral environment and the image turns to blue (C-C”). The result matched findings in which the progression of mitophagy causes mitochondria to be engulfed in lysosomes when in an acidic environment.
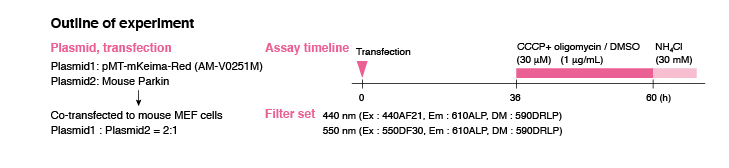
Ratio Imaging
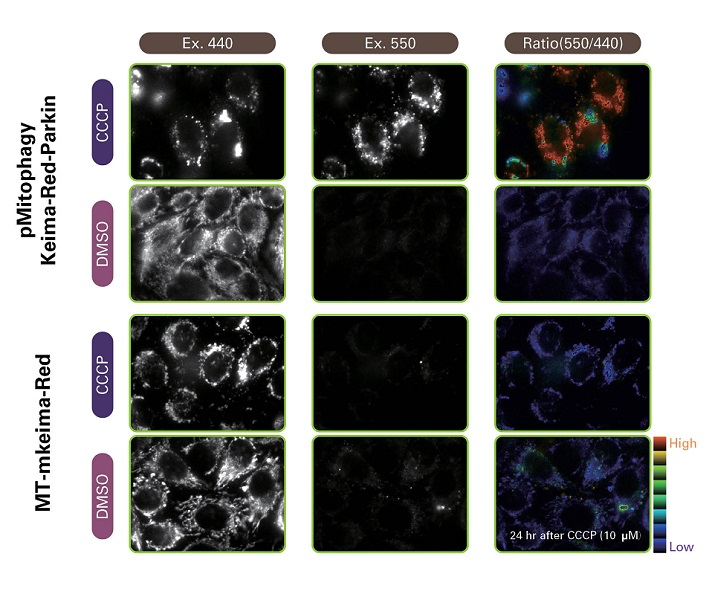
Mitophagy flux is analysed by the fluorescent intensity ratio of 440 nm and 550 nm. A strong signal at 440nm indicates that mitophagy did not occur while a strong signal at 550 nm indicates mitophagy occurred.
High Content Analysis by IN Cell Analyser
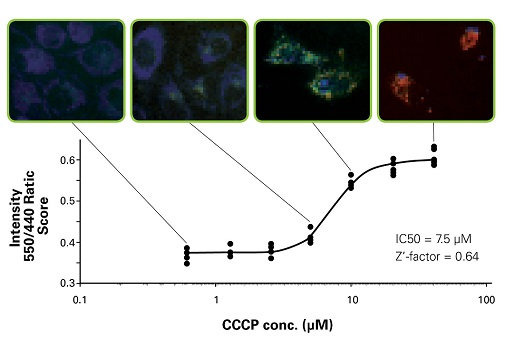
Mitophagy was detected using IN Cell Analyser 1000. MBL obtained a reasonable IC50 value and a sufficient Z’-factor. pMitophagy Keima-Red -Parkin provides a robust assay for mitophagy monitoring in living cells using high content analyser.
Originally posted on: https://resources.mblintl.com/learning-center/research-area/fluorescent-proteins/mitophagy-detecton/?hsCtaTracking=ae7f8052-09b5-4249-8ec4-cd2e69e38519%7Cf84dab52-c0b2-4ae9-962b-b77876a440c3
Caltag Medsystems is the distributor of MBL products in the UK and Ireland. If you have any questions about these products, please contact us.
